THE FUTURE OF THE FLU SHOT
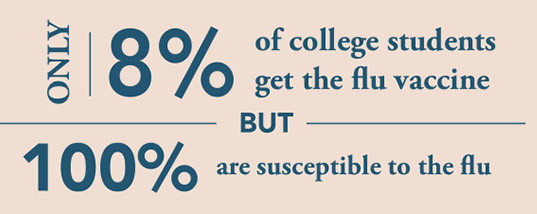
source: University of South Carolina Health Services
BY ANA MEHRANIAN – It all starts with a scratchy throat, a few sniffles, and an unexpected sneeze or two. Before you know it, you can barely roll out of bed, breathing through your nose is not an option, and class started thirty minutes ago. Most of us are familiar with this feeling and its culprit: the flu. Caused by a virus with an incubation period as short as twenty-four hours, the onset of flu symptoms appear too quickly to curb, yet often linger longer than expected. On college campuses, the influenza virus circulates rapidly due to the high volume of students sharing living and study spaces, as well as restrooms and classrooms. According to the Centers for Disease Control and Prevention (CDC), college students infected with the influenza virus experience symptoms for an average of eight or more days.
An extremely contagious respiratory illness, the flu is caused by influenza A, B, or C viruses. The virus itself is circular and covered in specialized glycoprotein “spikes” comprised of hemagglutinin and neuraminidase. These proteins allow the virus to enter and exit the host cell upon initiation and completion of the viral life cycle. Variants and combinations of these surface proteins (e.g. H1, H2, H3, N1, N2, N3, etc.) make each virus unique, altering the ability of each virus to spread and cause illness.
The influenza virus diversifies through genetic shift and drift, patterns of genetic change and recombination made possible by the chemical makeup of the virus’s genetic code: ribonucleic acid. During RNA synthesis and replication, incorrect nucleotides are often inserted and misincorporated into the new RNA chain. These copying errors are the driving force of viral evolution, with rapid mutations having the potential to cause pandemics. In other words, the flu is so successful because it makes mistakes. According to the World Health Association (WHO), there are 3-5 million cases of severe influenza every year, causing between 250,000 and 500,000 deaths annually.
After the influenza virus enters your body, it travels to the cells lining your respiratory tract and begins replication. To do this, the virus must first gain entry to its chosen host cell. The surface protein hemagluttanin acts as a key, unlocking the door to the cell membrane. Once the virus is safely inside the cell, it bursts, releasing its genetic sequence into the host cell’s nucleus. The host cell is then transformed into a factory for virus production and export. As the body detects these invaders, it recruits immune cells, sending them to the areas containing the replicating viruses. At this point, the immune system has been kicked into gear and is actively attacking viral cells to halt their proliferation.
How can we prevent all of this in the first place? The answer is in vaccination. Traditional seasonal flu shots are trivalent vaccines, meaning that they provide protect against three influenza viruses: an influenza A (H1N1) virus, an influenza A (H3N2) virus, and an influenza B virus. The specific types and strains are chosen every year based on the viruses research indicates as the most common for that season. Why not create a vaccine that protects against all possible combinations? The antigenic differences between influenza types A and B as well as subtype and strain diversity has caused difficulty in creating effective and all-encompassing influenza vaccinations. Within each influenza season and strain, there is an (almost) unpredictable combination of both influenza A and B lineages. This creates a “mismatch” between the vaccinations and circulating strains.
Dr. Ted Ross, a microbiologist and the director of the Center for Vaccines and Immunology at the University of Georgia (UGA), is attempting to face these challenges as he and his colleagues work to create a universal or broadly reactive influenza vaccine. The approach taken by Ross and his team involve combining mutations of the surface protein hemagluttanin from every flu strain to ever exist into a single molecule with the potential to build the foundation of a new vaccine. The data technique used in the process is called computationally optimized broadly reactive antigen (COBRA). COBRA employs computer algorithms to analyze genome sequence information and select antigens with the characteristics to represents a wide variety of clades. These antigens are then used to produce a vaccine with a wide cross-clade coverage and high potency. This immunological breadth is important to the unpredictable tendency of influenza to mutate and evolve.
In a study conducted by Ross’ laboratory, a COBRA-derived vaccine was designed to protect against all possible variations of the H1N1 virus. Mice treated with the vaccine were successful in developing the necessary antibodies and were granted immunity. The results were published in the Journal of Virology in 2016, and as of last year, the vaccine has been planned for testing in clinical trials. In the words of Dr. Ross, “The development of a universal or broadly reactive influenza vaccine will be a paradigm changing event in vaccinology. A vaccine that can protect all people, from infants to the elderly, pregnant women, the immunocompromised for 5, 10 or more years will change our lives.” The future for a universal flu shot is bright, and part of it is happening right here at UGA.

